This page discusses prostate cancer molecular testing, divided into three sections:
- ConfirmMDx
- Decipher
- PTEN and ERG Gene Mutations
ConFirmMDx
Overview
ConfirmMDx is a commercially available molecular test offered by MDxHealth that is used to aid in the detection of prostate cancer. It is an epigenetic assay that helps in identifying patients who may have false-negative prostate core biopsies. The test is typically performed on benign prostate biopsies to determine if the benign biopsies truly represent an absence of prostate cancer, or if there is molecular evidence suggesting the possibility of carcinoma in the unsampled tissue. The company claims that the ConfirmMDX test has a 90% negative predictive value.
Rationale
MDxHealth reports that up to 25% of patients may have false-negative prostate biopsies. Despite negative biopsies, these patients may be subjected to multiple repeat biopsies due to rising PSA levels. As an alternative to repeat biopsies based upon increasing PSA levels, MDxHealth offers a molecular analysis that can be performed following the patients’ initial benign biopsies. If the molecular analysis does not reveal any abnormalities suggestive of malignancy, the clinician can use the negative molecular assay result as evidence that the initial biopsy was a true negative, and subsequent repeat biopsies may not be necessary.
Alternatively, if the molecular analysis reveals abnormalities that suggest the possibility of an underlying carcinoma, then the patient can undergo immediate focused repeat biopsies based upon the mapping results of the molecular assay.
Methodology and Clinical Application
ConfirmMDx is a proprietary quantitative methylation-specific PCR test. Abnormal patterns of DNA methylation have been associated with various malignancies. In the prostate, tissue involved by as well as tissue nearby carcinoma has been found to show aberrant DNA methylation. Therefore, benign tissue adjacent to carcinoma may have an abnormal methylation pattern that can be detected by the assay. The test is intended to be performed on multiple-part biopsies which designate the specific location of each biopsy within the prostate. By identifying the anatomic areas which show abnormal methylation, the patient can undergo immediate repeat biopsies which focus specifically on the suspicious anatomic locations identified by the assay. This may help identify carcinoma earlier and without numerous repeat biopsies of the entire prostate.
Specimen Requirements
ConfirmMDX can be performed on formalin-fixed, paraffin-embedded (FFPE) tissue blocks or unstained slides. To order the test or for more information, please refer to the
following link http://mdxhealth.com/products-and-technology/products/how-to-order-confirmmdx.
ConfirmMDx is a commercially available molecular test offered by MDxHealth that is used to aid in the detection of prostate cancer. It is an epigenetic assay that helps in identifying patients who may have false-negative prostate core biopsies. The test is typically performed on benign prostate biopsies to determine if the benign biopsies truly represent an absence of prostate cancer, or if there is molecular evidence suggesting the possibility of carcinoma in the unsampled tissue. The company claims that the ConfirmMDX test has a 90% negative predictive value.
Rationale
MDxHealth reports that up to 25% of patients may have false-negative prostate biopsies. Despite negative biopsies, these patients may be subjected to multiple repeat biopsies due to rising PSA levels. As an alternative to repeat biopsies based upon increasing PSA levels, MDxHealth offers a molecular analysis that can be performed following the patients’ initial benign biopsies. If the molecular analysis does not reveal any abnormalities suggestive of malignancy, the clinician can use the negative molecular assay result as evidence that the initial biopsy was a true negative, and subsequent repeat biopsies may not be necessary.
Alternatively, if the molecular analysis reveals abnormalities that suggest the possibility of an underlying carcinoma, then the patient can undergo immediate focused repeat biopsies based upon the mapping results of the molecular assay.
Methodology and Clinical Application
ConfirmMDx is a proprietary quantitative methylation-specific PCR test. Abnormal patterns of DNA methylation have been associated with various malignancies. In the prostate, tissue involved by as well as tissue nearby carcinoma has been found to show aberrant DNA methylation. Therefore, benign tissue adjacent to carcinoma may have an abnormal methylation pattern that can be detected by the assay. The test is intended to be performed on multiple-part biopsies which designate the specific location of each biopsy within the prostate. By identifying the anatomic areas which show abnormal methylation, the patient can undergo immediate repeat biopsies which focus specifically on the suspicious anatomic locations identified by the assay. This may help identify carcinoma earlier and without numerous repeat biopsies of the entire prostate.
Specimen Requirements
ConfirmMDX can be performed on formalin-fixed, paraffin-embedded (FFPE) tissue blocks or unstained slides. To order the test or for more information, please refer to the
following link http://mdxhealth.com/products-and-technology/products/how-to-order-confirmmdx.

Decipher
Overview
Decipher is a commercially available molecular test offered by GenomeDx Biosciences that may help guide treatment decisions after prostatectomy in patients with prostate cancer. It is a genomic classifier test that is used to determine the risk of metastasis in patients following surgery. The test helps distinguish high-risk patients from low-risk patients, so that patients found to have low-risk genomic classifier results may be able to avoid radiation therapy after surgery. According to GenomeDx, the test can reclassify up to 60% of patients into a low-risk category, who were originally deemed high-risk by clinical and pathologic findings. Of these patients reclassified into a low-risk category by Decipher, 98.5% remained metastasis-free after five years.
Rationale
GenomeDx reports that up to 50% of patients who undergo surgery for prostate cancer are clinically considered to be at high-risk for metastatic disease, based on clinical parameters and pathology. Therefore, these patients are often treated with radiation therapy. However, 90% of these patients do not develop metastases or die from their disease. This finding provides some evidence that patients may be undergoing radiation therapy that does not significantly affect their risk of metastasis or overall survival. Furthermore, radiation therapy adds considerable cost and has multiple possible complications or adverse side effects, which may result in the need for additional treatment. Decipher provides an alternative method of determining prostate cancer aggressiveness, irrespective of Gleason score, PSA levels, or other clinical findings. Therefore, the test helps identify additional patients eligible to avoid radiation therapy, who without the molecular data may have received potentially unneeded treatment.
Methodology and Clinical Application
According to GenomeDx, the test was developed by initially analyzing the expression of over one million genes within the human genome. These genes were analyzed by proprietary genome-wide search algorithms. After this analysis, 22 genes were found to be most useful for predicting which prostate carcinomas will have an aggressive clinical course resulting in metastases and poor clinical outcomes. The results Decipher provides to the clinician are based upon the algorithmic analysis of these 22 genes. The test results provide an estimate of tumor aggressiveness that can be used to determine appropriate patient follow-up and the need for subsequent radiotherapy.
Specimen Requirements
Decipher can be performed on formalin-fixed, paraffin-embedded (FFPE) tissue blocks. To order the test or for more information, please refer to the following link http://genomedx.com/decipher-test/addressing-challenges-prostate-cancer/
Decipher is a commercially available molecular test offered by GenomeDx Biosciences that may help guide treatment decisions after prostatectomy in patients with prostate cancer. It is a genomic classifier test that is used to determine the risk of metastasis in patients following surgery. The test helps distinguish high-risk patients from low-risk patients, so that patients found to have low-risk genomic classifier results may be able to avoid radiation therapy after surgery. According to GenomeDx, the test can reclassify up to 60% of patients into a low-risk category, who were originally deemed high-risk by clinical and pathologic findings. Of these patients reclassified into a low-risk category by Decipher, 98.5% remained metastasis-free after five years.
Rationale
GenomeDx reports that up to 50% of patients who undergo surgery for prostate cancer are clinically considered to be at high-risk for metastatic disease, based on clinical parameters and pathology. Therefore, these patients are often treated with radiation therapy. However, 90% of these patients do not develop metastases or die from their disease. This finding provides some evidence that patients may be undergoing radiation therapy that does not significantly affect their risk of metastasis or overall survival. Furthermore, radiation therapy adds considerable cost and has multiple possible complications or adverse side effects, which may result in the need for additional treatment. Decipher provides an alternative method of determining prostate cancer aggressiveness, irrespective of Gleason score, PSA levels, or other clinical findings. Therefore, the test helps identify additional patients eligible to avoid radiation therapy, who without the molecular data may have received potentially unneeded treatment.
Methodology and Clinical Application
According to GenomeDx, the test was developed by initially analyzing the expression of over one million genes within the human genome. These genes were analyzed by proprietary genome-wide search algorithms. After this analysis, 22 genes were found to be most useful for predicting which prostate carcinomas will have an aggressive clinical course resulting in metastases and poor clinical outcomes. The results Decipher provides to the clinician are based upon the algorithmic analysis of these 22 genes. The test results provide an estimate of tumor aggressiveness that can be used to determine appropriate patient follow-up and the need for subsequent radiotherapy.
Specimen Requirements
Decipher can be performed on formalin-fixed, paraffin-embedded (FFPE) tissue blocks. To order the test or for more information, please refer to the following link http://genomedx.com/decipher-test/addressing-challenges-prostate-cancer/
PTEN and ERG Gene Mutations
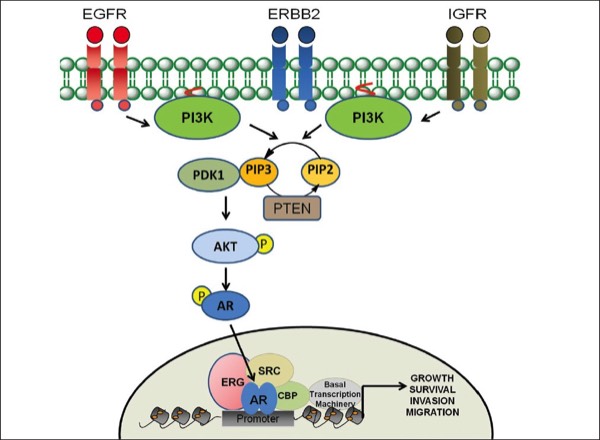
Diagram adapted from Dasgupta S et al. Oncogenic activation in prostate cancer progression and metastasis: Molecular insights and future challenges. J Carcinog. 2012;11:4.
Two known genetic alterations in prostate carcinoma that are associated with aggressive behavior and poor prognosis involve the PTEN and ERG genes.
PTEN is a tumor suppressor gene that may be deleted in up to 40% of prostate cancers. Hemizygous PTEN genomic deletions occur in approximately 39% of prostate cancers, and homozygous deletions are seen in approximately 5% of prostate cancers. PTEN loss has been associated with earlier onset of biochemical recurrence after surgery (as determined by serum PSA levels). It also correlates with more advanced disease at the time of surgery with features such as positive surgical margins, extraprostatic extension, and seminal vesicle involvement. Homozygous and hemizygous deletions have both been associated with more aggressive tumors with higher metastatic potential, when compared to prostate cancer patients with a normal PTEN profile. As compared with hemizygous deletions, homozygous deletions are associated with even more adverse clinical outcomes and much earlier biochemical relapse. Therefore, PTEN testing may be useful for both guiding patient management and determining prognosis.
ERG is an oncogene that functions as a transcriptional regulator. Prostate cancer patients with wild-type ERG have a favorable prognosis with a 90% 8-year survival rate. Patients with ERG deletion show a less favorable prognosis with a 75% 8-year survival rate. Patients with ERG deletion and polysomy have an extremely poor prognosis with a 25% survival rate. Therefore, detection of ERG gene abnormalities can be helpful as a prognostic indicator and for tailoring post-operative care by identifying patients with aggressive tumors.
Testing for both PTEN and ERG gene alterations can be done by FISH analysis on formalin-fixed, paraffin-embedded (FFPE) tissue blocks. Some of the companies that offer PTEN and/or ERG FISH analysis include PersonalizeDx, Pacific Diagnostics, and Neogenomics Laboratories.
https://www.personalizedx.com/prostate-cancer.html
http://www.pacificdx.com/test.html
http://www.neogenomics.com/index.htm
References
1. Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. British Journal of Cancer. 2007;97:678-685.
2. Reid AHM, Attard G, Ambroisine L, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. British Journal of Cancer. 2010;102(4):678-684.
3. Krohn A, Diedler T, Burkhardt L, et al. Genomic Deletion of PTEN Is Associated with Tumor Progression and Early PSA Recurrence in ERG Fusion-Positive and Fusion-Negative Prostate Cancer. The American Journal of Pathology. 2012;181(2):401–412.
4. Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene 2008;27:253–263.
5. Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS-ERG Gene Fusion is Not Associated with Outcome in Patients Treated by Prostatectomy. Cancer Research. 2009;69(4):1400-1406.
6. Toubaji A, Albadine R, Meeker AK, et al. Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas. Modern Pathology. 2011;24:1511-1520.
7. Dasgupta S, Srinidhi S, Vishwanatha JK. Oncogenic activation in prostate cancer progression and metastasis: Molecular insights and future challenges. Journal of Carcinogenesis. 2012;11:4.

